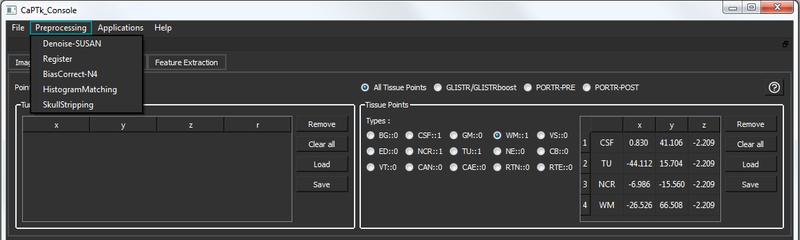
An essential component for quantitative image analysis is the appropriate pre-processing steps. The related CaPTk tools available for this purpose are fully-parameterizable and comprise:
- Denoising. Intensity noise reduction in regions of uniform intensity profile is offered through a low-level image processing method, namely Smallest Univalue Segment Assimilating Nucleus (SUSAN) [1].
- Bias correction. Correction for magnetic field inhomogeneity is provided using a non-parametric non-uniform intensity normalization [2].
- Co-registration. Registration of various images to the same anatomical template, for examining anatomically aligned imaging signals in tandem and at the voxel level, is offered by the ITK module ITKRegistrationCommon (i.e., a combination of itk::MattesMutualInformationImageToImageMetric, itk::RegularStepGradientDescentOptimizerv4, and itk::MultiResolutionImageRegistrationMethod).
- Skull-stripping. Removing the bone structure in brain scans is performed using the ITK filter itk::StripTsImageFilter [3].
- Intensity normalization. Conversion of signals across modalities are converted to comparable quantities using histogram matching [4].
- DICOM Conversion. Dicom to NIfTI conversion using dcm2nii. Please move DICOM series to individual directories for this to work. DTI and Perfusion images are currently unsupported [7].
- DTI-FA (Fractional Anisotropy)
- DTI-AX (Axial Diffusivity)
- DTI-RAD (Radial Diffusivity)
- DTI-ADC (Apparent Diffusion Coefficient)
- DSC-rCBV (relative Cerebral Blood Volume)
- DSC-PH (Peak Height)
- DSC-PSR (Percentage Signal Recovery)
All these tools can be found under the menu option: 'pre-processing'.
References:
[1] S.M.Smith, J.M.Brady, "SUSAN - a new approach to low level image processing", International Journal of Computer Vision 23, 45–78, 1997
[2] N.J.Tustison, B.B.Avants, P.A.Cook, Y.Zheng, A.Egan, P.A.Yushkevich, J.C.Gee, "N4ITK: Improved N3 Bias Correction", IEEE Transactions on Medical Imaging 29, 1310-1320, 2010
[3] S.Bauer, L.P.Nolte, M.Reyes, "Skull-stripping for Tumor-bearing Brain Images", In Annual Meeting of the Swiss Society for Biomedical Engineering, page 2, Bern, 2011
[4] L.G.Nyul, J.K.Udupa, X.Zhang, "New Variants of a Method of MRI Scale Standardization", IEEE Transactions on Medical Imaging, 19(2):143-150, 2000
[5] J.M.Soares, P.Marques, V.Alves, N.Sousa. "A hitchhiker's guide to diffusion tensor imaging", Frontiers in neuroscience, 7, 2013
[6] E.S.Paulson, K.M.Schmainda, "Comparison of dynamic susceptibility-weighted contrast-enhanced MR methods: recommendations for measuring relative cerebral blood volume in brain tumors", Radiology, 249(2):601-613, 2008 [7] MRIcroGL: https://www.nitrc.org/projects/mricrogl/
