CaPTk is developed and maintained by the Center for Biomedical Image Computing and Analytics (CBICA) at the University of Pennsylvania, and draws upon research from several groups within the Center.
New applications, written in any programming language, can be integrated into CaPTk at different levels. These applications can then run within CaPTk, while having direct access to the full breadth of CaPTk’s interactive capabilities.
- Source level integration: At this level, the new application source code (C++) is compiled alongside CaPTk, ensuring the most optimized integration. Source-level integration is straight-forward (only requiring additions to relevant CMake files and minor additions to the interactive base) if the new application relies on a subset of CaPTk’s dependencies (i.e., ITK, VTK, OpenCV, Qt).
- Executable level integration: This level provides a graphical interface to an existing command-line application (not necessarily developed in C++), allowing users to leverage CaPTk’s functionality (e.g., interaction, feature extraction). Executable-level integration requires only minor additions to CaPTk to create a menu option for the new application.
Almost every application of CaPTk has an accompanying command-line executable (with more on the way). Those programs can be called directly, making the CaPTk applications available as components within a larger pipeline or for efficient batch processing of large numbers of images.
We will provide the technical details of the Cancer Imaging Phenomics Toolkit (CaPTk) using which new applications can be integrated into the global framework and also optimize/improve the code. For any questions/details, please feel free to email CBICA Software.
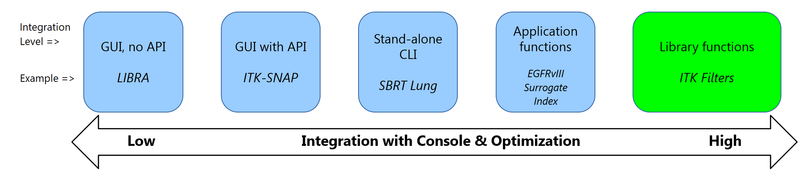
- LIBRA, in its current form (MATLAB executable) has the least possible integration with the CaPTk Console; the console can call the executable, which launches its own UI for the user.
- ITK-SNAP is an example of integration where CaPTk Console communicates with the application using the provided API; in this case CaPTk ensures that the loaded images, ROIs, masks, etc. are all passed to ITK-SNAP and the result from the segmentation step there gets passed to the console for visualization after ITK-SNAP is closed.
- SBRT Lung is an example of integration with a stand-alone CLI application. The Console calls the executable in accordance with CLI, passes the loaded images and ROIs, etc. and visualizes the result when the application is done.
- EGFRvIII Surrogate Index provides the tightest possible integration with the CaPTk Console. All data (loaded images, ROIs, etc.) is passed in-memory and the visualization of results happens instantaneously.
- All functions available in native C++ libraries (ITK, OpenCV, VTK) are available for native applications to use.
Dependencies
-
The Graphical Layer is currently written on a Qt4 based framework on C++ for speed, stability and extensibility. Qt, thus, becomes the first dependency for compiling CaPTk. Qt was chosen because it is a well-known tool for developing GUI applications both in academia and also in industry.
-
The basic file input/output operations are based on standard ITK I/O, thereby making it the second dependency. ITK was chosen on account of it being an industry and academic standard for developing medical image applications. It also has one of the most vibrant developer and user communities. Currently supported data formats are DICOM and NIFTI.
-
Rendering the data is done using VTK, making it the third dependency. VTK has been specifically designed for medical image data and it utilizes various hardware rendering techniques which make applications developed on it very fluid.
-
CMake is used to configure the project. This is the industry standard for cross-platform compilation.
- OpenCV
- A C++ compiler (we develop on MSVC/2013 and GCC/4.9.2).
Integrating your C++ application into CaPTk
Let’s say the name of your application is yourSourceApp. The following steps highlight the steps required for you to integrate your application into CaPTk:
- Input and Output of files is handled by the graphical layer, which takes into account file handling, directory sorting, etc. so you can worry about the important stuff, i.e., your algorithm.
- We are currently developing actively on MSVC/12 - Visual Studio 2013 and GCC/4.9.2. Plans are in motion to migrate to MSVC/14 - Visual Studio 2015 and GCC/5.x very soon, upon which all C++11 features will be enabled by default.
- The graphical layer reads DICOM or NIfTI files and passes it on as an ITK-Image. The data type defaults to the same type in which the original image was written. This is done using the ITK-ImageIOBase class.
- Your application should read either a single ITK-Image or a vector of ITK-Images and give output in a similar format (either a single ITK-Image or a vector of ITK-Images).
- yourSourceApp should be structured as a single class, i.e., a collection of yourSourceApp.h and yourSourceApp.cpp (ensure that the extensions match up otherwise CMake won’t pick them up as valid applications). Put the class implementation in the following folder - $PROJECT_SOURCE_DIR/src/applications and let CMake pick your application up in the next compilation step.
- If your algorithm does any kind of compilation or builds dependencies (libraries, executables, etc.), ensure that all the new files (non-source code files) are generated out-of-source. This is done to ensure that packaging the final application can happen in a concurrent manner.
-
CaPTk is able to handle application-specific dependencies well. For example, if you prefer the SVM implementation of LibSVM rather than that of OpenCV, you can simply create a new folder called /yourSourceApp_includes under $PROJECT_SOURCE_DIR/src/applications and let CMake take care of the configuration. yourSourceApp will see the includes as if it was present in the same folder (i.e., no need to specify folder when including these dependencies).
Integrating your Python application into CaPTk
Let’s say the name of your application is yourPythonApp. The following steps give a brief high-level overview regarding the steps required for integrating it with CaPTk:
- Input and Output of files is handled by the graphical layer, which takes into account file handling, directory sorting, etc. so you can worry about the important stuff, i.e., your algorithm.
- For interpreted languages such as Python, the graphical layer writes a NIfTI file (yes, there is disk I/O, yes it is inefficient, yes it is stupid but there is no other way around it so just deal with it).
- Your application should be structured as a single yourPythonApp.py script (can be a class or pipeline). This should be present in the $PROJECT_SOURCE_DIR/src/applications/py directory. Your application should also have a file analogous to the setup.py file used in Python projects by the name yourPythonApp_setup.py.
- The Python configuration creates a virtual environment in the $PROJECT_BINARY_DIR/py directory, where all the dependencies are constructed.
- Your application should support command line interfaces properly (verbose parameters throughout at the very least).
-
Add a new application under the APP_LIST_PY_GUI in
CaPTk_source_code/src/applications/CMakeLists.txtand then make the corresponding addition in thefMainWindowandui_fMainWindowclass (take LIBRA as a starting point). -
Once you have a menu item and the corresponding function call for your application, you can recompile CaPTk and then it will be able to pick up your application
